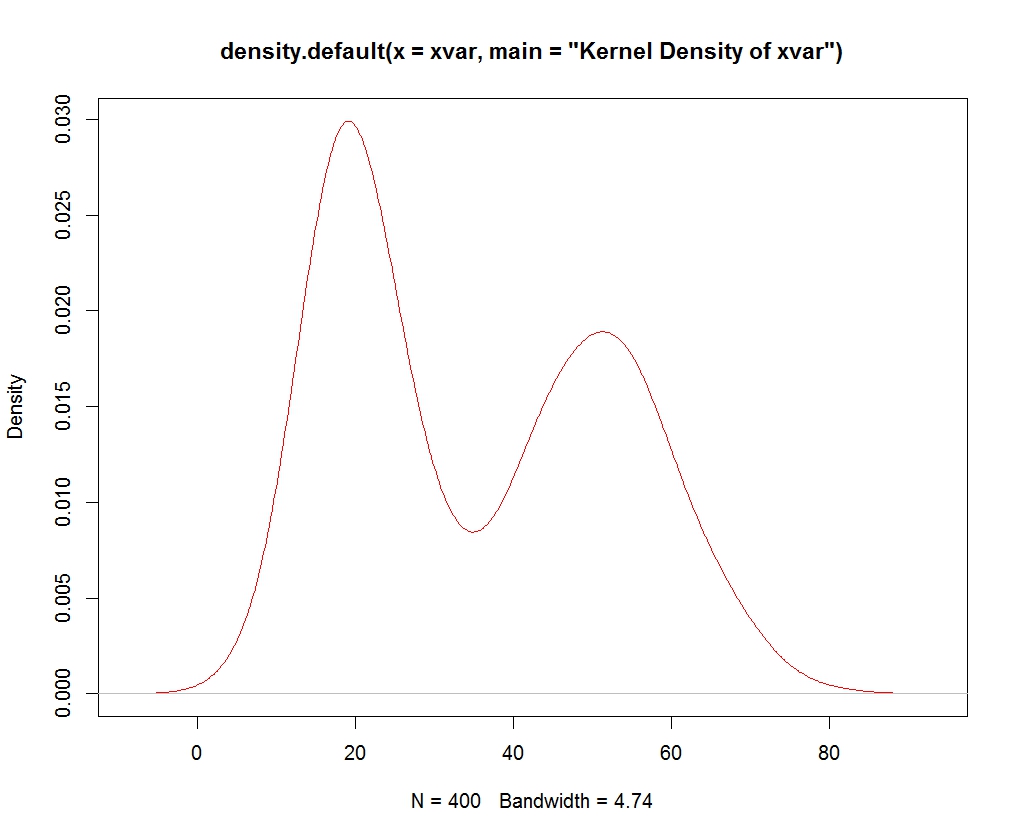
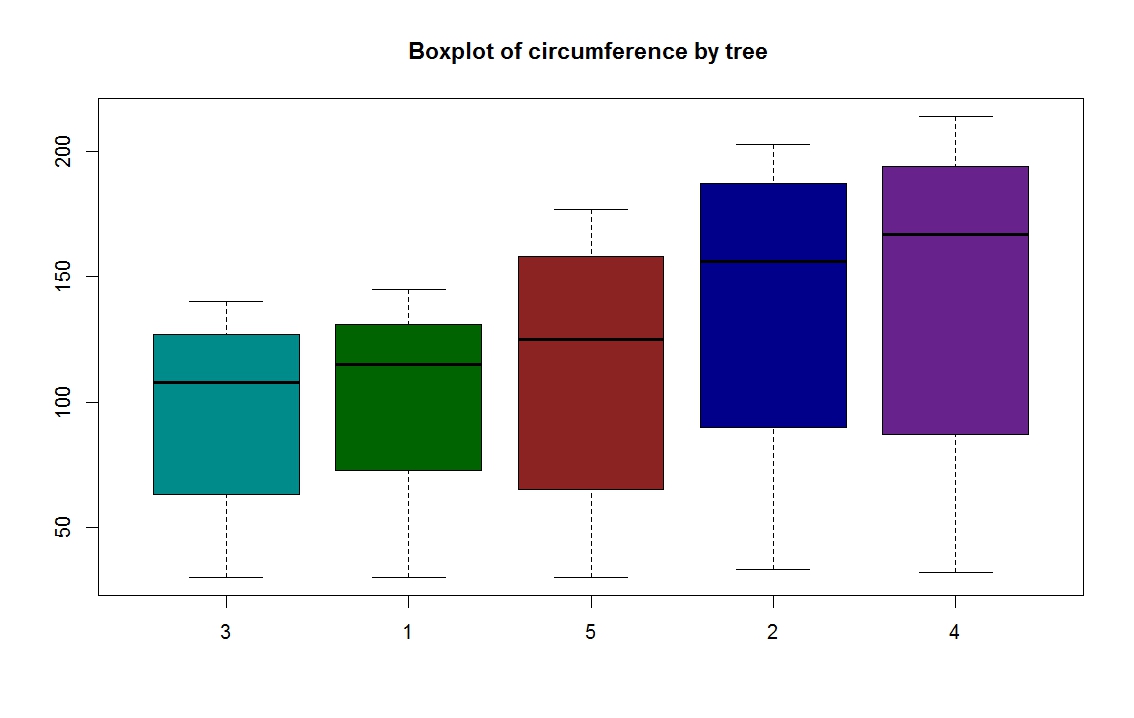
set.seed(13)
x <- c(rnorm(1000, 50, 20), rnorm(500, 70, 10), rnorm(500, 30, 10), runif(1000, 0, 100))
g <- rep (c("normal", "binormal", "uniform"), each = 1000)
# default trellis box plot
require(lattice)
#regular box plot
bwplot(g ~ x, col = "red", fill = c("cyan4", "tan", "blue"))
# box-percentile plot with data density (rug plot)
bwplot(g ~ x, panel=panel.bpplot, probs=seq(.01,.49,by=.01), datadensity=TRUE,
col = "red" )
# continuous plot up until quartiles ("Tootsie Roll plot")
bwplot(g ~ x, panel=panel.bpplot, probs=seq(.01,.25,by=.01))
# start at quartiles then make it continuous ("coffin plot")
bwplot(g ~ x, panel=panel.bpplot, probs=seq(.25,.49,by=.01))
# same as previous but add a spike to give 0.95 interval
bwplot(g ~ x, panel=panel.bpplot, probs=c(.025,seq(.25,.49,by=.01)))
# decile plot with reference lines at outer quintiles and median
bwplot(g ~ x, panel=panel.bpplot, probs=c(.1,.2,.3,.4), qref=c(.5,.2,.8))
# prototype of bplot
bpplt()