The blog is a collection of script examples with example data and output plots. R produce excellent quality graphs for data analysis, science and business presentation, publications and other purposes. Self-help codes and examples are provided. Enjoy nice graphs !!

Labels
2d
(1)
3 vartiable plots
(5)
3D plots
(8)
arch
(1)
area
(1)
association plot
(4)
bar
(1)
barchart
(13)
bean plot
(1)
beeswarm
(1)
binormial
(1)
biplot
(1)
box-percentile
(2)
box-whisker plot
(1)
boxplot
(10)
bubble plot
(5)
calendar
(1)
categorical data
(6)
centepede plot
(1)
circle
(2)
circular
(1)
cluster
(4)
color
(2)
colour
(1)
combination plot
(10)
countur
(1)
cross bar
(1)
cumulative
(1)
curve
(3)
dendogram
(3)
density
(13)
diagram
(2)
distribution
(9)
ditribution
(1)
dot plot
(1)
double axis
(1)
ellipse
(2)
error bar
(6)
factor plot
(3)
fluctutation diagram
(1)
google
(1)
grid plot
(1)
heatmap
(20)
hexabin plot
(1)
histogram
(11)
hive
(1)
kernel density
(4)
ladder plot
(2)
large data points
(4)
level plot
(1)
line plot
(3)
line range
(1)
manhattan plot
(1)
map
(13)
mosaic plot
(1)
normal
(2)
notched
(1)
parallel plot
(1)
pedigree plot
(1)
phylogentic tree
(1)
piechart
(3)
points
(2)
polar
(1)
Q-Q plot
(1)
raster
(2)
regression line
(3)
ribbon plot
(1)
rootogram
(1)
rugs
(2)
scale plot
(1)
scenes
(1)
shaded
(1)
spatial plot
(2)
sphere
(1)
spike histogram
(1)
Spine plot
(1)
stacked bar
(1)
Sunflower
(1)
ternary plot
(1)
text only
(1)
timeseries
(6)
trellis plot
(8)
two axis
(1)
vinn diagram
(1)
voilin plot
(2)
wireframe plot
(1)
xy barplot
(4)
xy line
(10)
xy points
(25)
Showing posts with label density. Show all posts
Showing posts with label density. Show all posts
Thursday, May 2, 2013
Sunday, April 28, 2013
RG#93: Add countour or heat map plot to XY scatter plot
# data
set.seed(1234)
n <- 10000
X = rnorm (n, 10, 4)
Y = X*1.5 + rnorm (n, 0, 8)
## colour brewing
library(RColorBrewer)
g = 11
my.cols <- rev(brewer.pal(g, "RdYlBu"))
#compute 2D kernel density
# kernel density using MASS
library(MASS)
z <- kde2d(X, Y, n=50)
plot(X, Y, xlab="X", ylab="Y", pch=19, cex=.3, col = "gray60")
contour(z, drawlabels=FALSE, nlevels=g, col=my.cols, add=TRUE, lwd = 2)
abline(h=mean(Y), v=mean(X), lwd=2, col = "black")
legend("topleft", paste("r=", round(cor(X, Y),2)), bty="n")
## estimate the z counts
prob <- c(.99, .95, .90, .8, .5, .1, 0.05)
dx <- diff(z$x[1:2])
dy <- diff(z$y[1:2])
sz <- sort(z$z)
c1 <- cumsum(sz) * dx * dy
levels <- sapply(prob, function(x) {
approx(c1, sz, xout = 1 - x)$y })
plot(X,Y, col = "gray80", pch = 19, cex = 0.3)
contour(z, levels= round (levels,7), add=T, col = "red")
# smooth scatter
require(KernSmooth)
smoothScatter(X, Y, nrpoints=.3*n, colramp=colorRampPalette(my.cols), pch=19, cex=.3, col = "green1")

set.seed(1234)
n <- 10000
X = rnorm (n, 10, 4)
Y = X*1.5 + rnorm (n, 0, 8)
## colour brewing
library(RColorBrewer)
g = 11
my.cols <- rev(brewer.pal(g, "RdYlBu"))
#compute 2D kernel density
# kernel density using MASS
library(MASS)
z <- kde2d(X, Y, n=50)
plot(X, Y, xlab="X", ylab="Y", pch=19, cex=.3, col = "gray60")
contour(z, drawlabels=FALSE, nlevels=g, col=my.cols, add=TRUE, lwd = 2)
abline(h=mean(Y), v=mean(X), lwd=2, col = "black")
legend("topleft", paste("r=", round(cor(X, Y),2)), bty="n")
## estimate the z counts
prob <- c(.99, .95, .90, .8, .5, .1, 0.05)
dx <- diff(z$x[1:2])
dy <- diff(z$y[1:2])
sz <- sort(z$z)
c1 <- cumsum(sz) * dx * dy
levels <- sapply(prob, function(x) {
approx(c1, sz, xout = 1 - x)$y })
plot(X,Y, col = "gray80", pch = 19, cex = 0.3)
contour(z, levels= round (levels,7), add=T, col = "red")
# smooth scatter
require(KernSmooth)
smoothScatter(X, Y, nrpoints=.3*n, colramp=colorRampPalette(my.cols), pch=19, cex=.3, col = "green1")

Thursday, April 18, 2013
RG#80: Plotting boxplot and histogram (overlayed or in margin)
# data
set.seed(4566)
data <- rnorm(100)
# layout where the boxplot is at top
nf <- layout(mat = matrix(c(1,2),2,1, byrow=TRUE), height = c(1,3))
par(mar=c(3.1, 3.1, 1.1, 2.1))
boxplot(data, horizontal=TRUE, outline=TRUE,ylim=c(-4,4), frame=F, col = "green1")
hist(data,xlim=c(-4,4), col = "pink")
# layout boxplot is at the bottom
nf <- layout(mat = matrix(c(1,2),2,1, byrow=TRUE), height = c(3,1))
par(mar=c(3.1, 3.1, 1.1, 2.1))
hist(data,xlim=c(-4,4), col = "pink")
boxplot(data, horizontal=TRUE, outline=TRUE,ylim=c(-4,4), frame=F, col = "green1", width = 10)
# Added to the plot:
par(mar=c(3.1, 3.1, 1.1, 2.1))
hist(data,xlim=c(-4,4), col = "pink")
boxplot(data, horizontal=TRUE, outline=TRUE, ylim=c(-4,4), frame=F, col = "green1", add = TRUE)
set.seed(4566)
data <- rnorm(100)
# layout where the boxplot is at top
nf <- layout(mat = matrix(c(1,2),2,1, byrow=TRUE), height = c(1,3))
par(mar=c(3.1, 3.1, 1.1, 2.1))
boxplot(data, horizontal=TRUE, outline=TRUE,ylim=c(-4,4), frame=F, col = "green1")
hist(data,xlim=c(-4,4), col = "pink")
# layout boxplot is at the bottom
nf <- layout(mat = matrix(c(1,2),2,1, byrow=TRUE), height = c(3,1))
par(mar=c(3.1, 3.1, 1.1, 2.1))
hist(data,xlim=c(-4,4), col = "pink")
boxplot(data, horizontal=TRUE, outline=TRUE,ylim=c(-4,4), frame=F, col = "green1", width = 10)
# Added to the plot:
par(mar=c(3.1, 3.1, 1.1, 2.1))
hist(data,xlim=c(-4,4), col = "pink")
boxplot(data, horizontal=TRUE, outline=TRUE, ylim=c(-4,4), frame=F, col = "green1", add = TRUE)
Wednesday, April 17, 2013
RG#77: Histogram and Cumulative Histogram with overlayed density plot
## Make some sample data
x <- sample(0:30, 200, replace=T, prob=15 - abs(15 - 0:30))
## Calculate and plot the two histograms
hcum <- h <- hist(x, plot=FALSE)
hcum$counts <- cumsum(hcum$counts)
plot(hcum, main="")
plot(h, add=T, col="grey")
## Plot the density and cumulative density
d <- density(x)
lines(x = d$x, y = d$y * length(x) * diff(h$breaks)[1], lwd = 2)
lines(x = d$x, y = cumsum(d$y)/max(cumsum(d$y)) * length(x), lwd = 2)
Labels:
combination plot,
cumulative,
density,
histogram
Tuesday, April 16, 2013
RG#68: Quantile comparison plot - QQ Plot (normal, t or F)
RG#67: Histogram with heatmap color in bars
# data set.seed(1234)
XV <- data.frame (x = rnorm(2000, 15, 4))
#plot
plot1 <- ggplot(data = XV, aes(x = x, y = ..density..)) +
geom_histogram(aes(fill = ..density..), binwidth = 0.25) +
stat_density(geom = "path", size = 1, alpha = 0.5) +
scale_y_continuous(expand = c(0.01, 0)) + scale_fill_gradient(low = "green4", high = "red") + xlab("Time") + ylab("Density") + theme_bw(16, "serif") +
theme(legend.position = "none")
print(plot1)
Friday, April 12, 2013
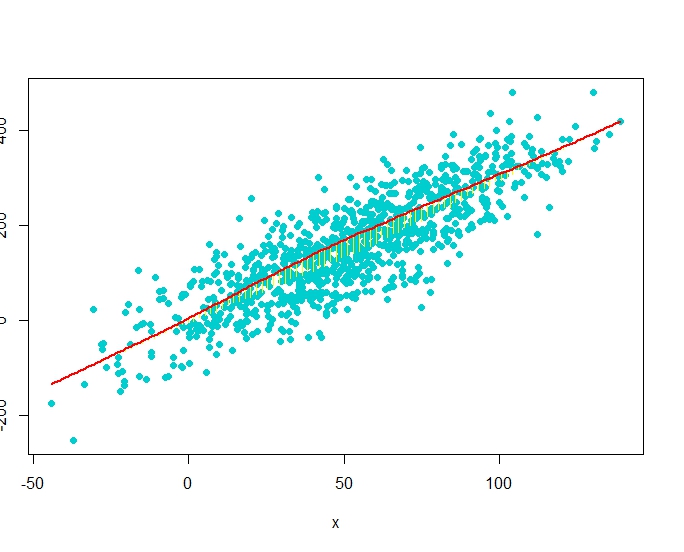
RG#54: Scatter Diagram with Rugs, Spike Histogram, or Density
# data
set(123)
x <- rnorm(1000, 50, 30)
y <- 3*x + rnorm(1000, 0, 20)
require(Hmisc)
plot(x,y)
#scat1d adds tick marks (bar codes. rug plot)
# on any of the four sides of an existing plot,
# corresponding with non-missing values of a vector x.
scat1d(x, col = "red") # density bars on top of graph
scat1d(y, 4, col = "blue") # density bars at right
plot(x,y, pch = 19)
histSpike(x, add=TRUE, col = "green4", lwd = 2)
histSpike(y, 4, add=TRUE,col = "blue", lwd = 2 )
histSpike(x, type='density',col = "red", add=TRUE) # smooth density at bottom
histSpike(y, 4, type='density', col = "red", add=TRUE)
plot(x,y, pch = 19)
smooth <- lowess(x, y) # add nonparametric regression curve lines(smooth, col = "red") # Note: plsmo() does this
scat1d(x, y=approx(smooth, xout=x)$y, col = "yellow") # data density on curve
scat1d(x, curve=smooth, col = "yellow") # same effect as previous command
# dataset(123)x <- rnorm(1000, 50, 30)y <- 3*x + rnorm(1000, 0, 60)
plot(x,y, pch = 19, col = "cyan3")
histSpike(x, curve=smooth, add=TRUE, col = "yellow") # same as previous but with histogram histSpike(x, curve=smooth, type='density', col = "red",lwd=2, add=TRUE)
Labels:
density,
histogram,
rugs,
spike histogram,
xy points
Tuesday, April 9, 2013
RG#49 : Sunflower plot
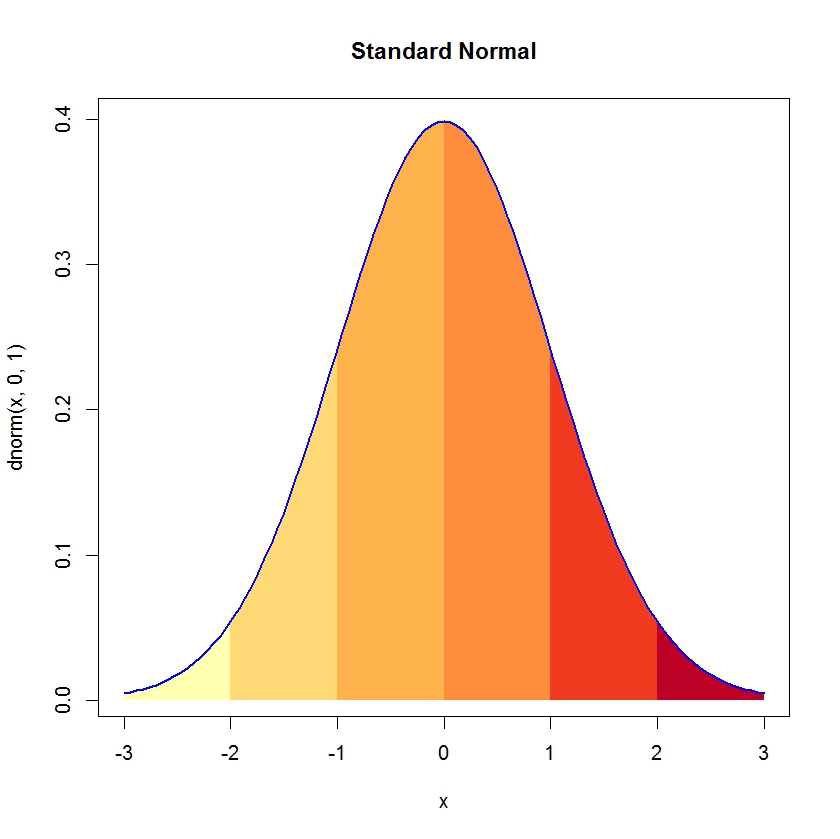
RG#47: shaded normal curve
codx <- c(-3,seq(-3,-2,0.01),-2)
cody <- c(0,dnorm(seq(-3,-2,0.01)),0)
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
polygon(codx,cody,col='red')
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
polygon(codx,cody, density = c(10, 20), angle = c(-45, 45))
# shading multiple polygons
cord.x1 <- c(-3,seq(-3,-2,0.01),-2)
cord.y1 <- c(0,dnorm(seq(-3,-2,0.01)),0)
cord.x2 <- c(-2,seq(-2,-1,0.01),-1)
cord.y2 <- c(0,dnorm(seq(-2,-1,0.01)),0)
cord.x3 <- c(-1,seq(-1,0,0.01),0)
cord.y3 <- c(0,dnorm(seq(-1,0,0.01)),0)
cord.x4 <- c(0,seq(0,1,0.01),1)
cord.y4 <- c(0,dnorm(seq(0,1,0.01)),0)
cord.x5 <- c(1,seq(1,2,0.01),2)
cord.y5 <- c(0,dnorm(seq(1,2,0.01)),0)
cord.x6 <- c(2,seq(2,3,0.01),3)
cord.y6 <- c(0,dnorm(seq(2,3,0.01)),0)
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
# brewing color for continious color filling
library(RColorBrewer)
plotclr <- brewer.pal(6,"YlOrRd")#
polygon(cord.x1,cord.y1,col= plotclr[1] , border = NA )
polygon(cord.x2,cord.y2,col=plotclr[2], border = NA )
polygon(cord.x3,cord.y3,col=plotclr[3], border = NA )
polygon(cord.x4,cord.y4,col=plotclr[4], border = NA )
polygon(cord.x5,cord.y5,col=plotclr[5], border = NA )
polygon(cord.x6,cord.y6,col=plotclr[6], border = NA )
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal', col = "blue", lwd = 2, add = TRUE)
# totally shaded curve
x <- seq(from=-3, to=3,by=0.001)
gb_gradient <-colorRampPalette(c("blue", "lightgreen", "red"))
cols <- gb_gradient (length(x))
curve(dnorm(x,0,1), xlim=c(-3,3))
segments(x, rep(0,length(x)),x,dnorm(x,0,1) , col=cols, lwd=2)
curve(dnorm(x,0,1), xlim=c(-3,3), col = "red", lwd=2, add = TRUE)
cody <- c(0,dnorm(seq(-3,-2,0.01)),0)
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
polygon(codx,cody,col='red')
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
polygon(codx,cody, density = c(10, 20), angle = c(-45, 45))
# shading multiple polygons
cord.x1 <- c(-3,seq(-3,-2,0.01),-2)
cord.y1 <- c(0,dnorm(seq(-3,-2,0.01)),0)
cord.x2 <- c(-2,seq(-2,-1,0.01),-1)
cord.y2 <- c(0,dnorm(seq(-2,-1,0.01)),0)
cord.x3 <- c(-1,seq(-1,0,0.01),0)
cord.y3 <- c(0,dnorm(seq(-1,0,0.01)),0)
cord.x4 <- c(0,seq(0,1,0.01),1)
cord.y4 <- c(0,dnorm(seq(0,1,0.01)),0)
cord.x5 <- c(1,seq(1,2,0.01),2)
cord.y5 <- c(0,dnorm(seq(1,2,0.01)),0)
cord.x6 <- c(2,seq(2,3,0.01),3)
cord.y6 <- c(0,dnorm(seq(2,3,0.01)),0)
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal')
# brewing color for continious color filling
library(RColorBrewer)
plotclr <- brewer.pal(6,"YlOrRd")#
polygon(cord.x1,cord.y1,col= plotclr[1] , border = NA )
polygon(cord.x2,cord.y2,col=plotclr[2], border = NA )
polygon(cord.x3,cord.y3,col=plotclr[3], border = NA )
polygon(cord.x4,cord.y4,col=plotclr[4], border = NA )
polygon(cord.x5,cord.y5,col=plotclr[5], border = NA )
polygon(cord.x6,cord.y6,col=plotclr[6], border = NA )
curve(dnorm(x,0,1),xlim=c(-3,3),main='Standard Normal', col = "blue", lwd = 2, add = TRUE)
# totally shaded curve
x <- seq(from=-3, to=3,by=0.001)
gb_gradient <-colorRampPalette(c("blue", "lightgreen", "red"))
cols <- gb_gradient (length(x))
curve(dnorm(x,0,1), xlim=c(-3,3))
segments(x, rep(0,length(x)),x,dnorm(x,0,1) , col=cols, lwd=2)
curve(dnorm(x,0,1), xlim=c(-3,3), col = "red", lwd=2, add = TRUE)
Monday, April 8, 2013
RG#35: density or Kernel density plot
Friday, April 5, 2013
RG#11:multiple histograms with normal distribution or density curve overlayed
#data
set.seed(1233)
data1 < - data.frame(pop =c(rep("A x B", 200), rep("A x C", 200), rep("B x C", 200) ) , var1 = c(rnorm(1000, 90,10), rnorm(1000, 50, 10), rnorm(1000, 20, 30)))
#plot
require(lattice)
# plot overall distribution
histogram(~ var1, data= data1, nint = 12, xlab = "trait1(measuring unit)", type = "density", panel = function(x, ...) {
panel.histogram(x, ...)
panel.mathdensity(dmath = dnorm, col = "black",
args = list(mean=mean(x),sd=sd(x)))
} )
# plot by each group
histogram(~ var1|factor(pop), data= data1, nint = 10, xlab = "trait1(measuring unit)", type = "density",
panel = function(x, ...) {
panel.histogram(x, col = "darkgreen", ...)
panel.mathdensity(dmath = dnorm, col = "red",
args = list(mean=mean(x),sd=sd(x)))
} )
set.seed(1233)
data1 < - data.frame(pop =c(rep("A x B", 200), rep("A x C", 200), rep("B x C", 200) ) , var1 = c(rnorm(1000, 90,10), rnorm(1000, 50, 10), rnorm(1000, 20, 30)))
#plot
require(lattice)
# plot overall distribution
histogram(~ var1, data= data1, nint = 12, xlab = "trait1(measuring unit)", type = "density", panel = function(x, ...) {
panel.histogram(x, ...)
panel.mathdensity(dmath = dnorm, col = "black",
args = list(mean=mean(x),sd=sd(x)))
} )
# plot by each group
histogram(~ var1|factor(pop), data= data1, nint = 10, xlab = "trait1(measuring unit)", type = "density",
panel = function(x, ...) {
panel.histogram(x, col = "darkgreen", ...)
panel.mathdensity(dmath = dnorm, col = "red",
args = list(mean=mean(x),sd=sd(x)))
} )
Labels:
density,
ditribution,
histogram,
kernel density
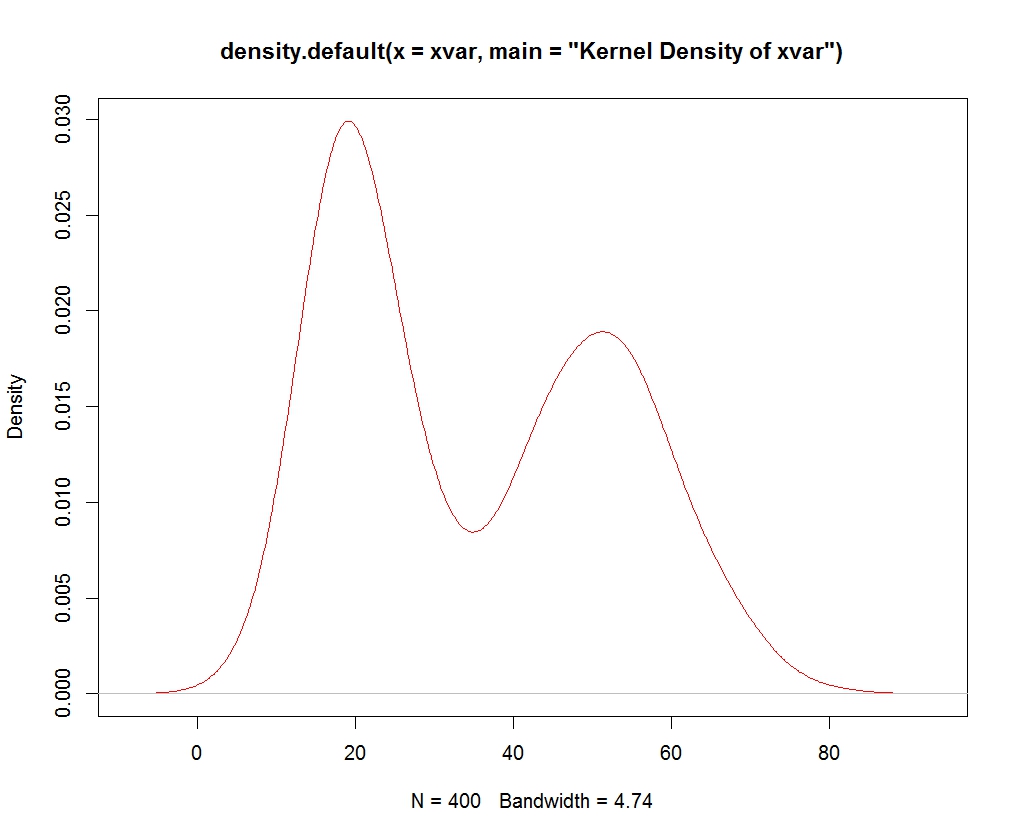
RG10 # plotting multiple suprimposed histograms or density plots
#data
set.seed(1233)
data1 <- data.frame(pop =c(rep("A x B", 200), rep("A x C", 200), rep("B x C", 200) ) , var1 = c(rnorm(1000, 90,10), rnorm(1000, 50, 10), rnorm(1000, 20, 30)))
#
plot using ggplot2
require(ggplot2)
qplot( var1, data = data1, geom = "density" , group = pop, fill = pop, alpha=.3) + theme_bw( )
qplot( var1, data = data1, geom = "histogram" , group = pop, fill = pop, alpha=.3) + theme_bw( )
set.seed(1233)
data1 <- data.frame(pop =c(rep("A x B", 200), rep("A x C", 200), rep("B x C", 200) ) , var1 = c(rnorm(1000, 90,10), rnorm(1000, 50, 10), rnorm(1000, 20, 30)))
#
plot using ggplot2
require(ggplot2)
qplot( var1, data = data1, geom = "density" , group = pop, fill = pop, alpha=.3) + theme_bw( )
qplot( var1, data = data1, geom = "histogram" , group = pop, fill = pop, alpha=.3) + theme_bw( )
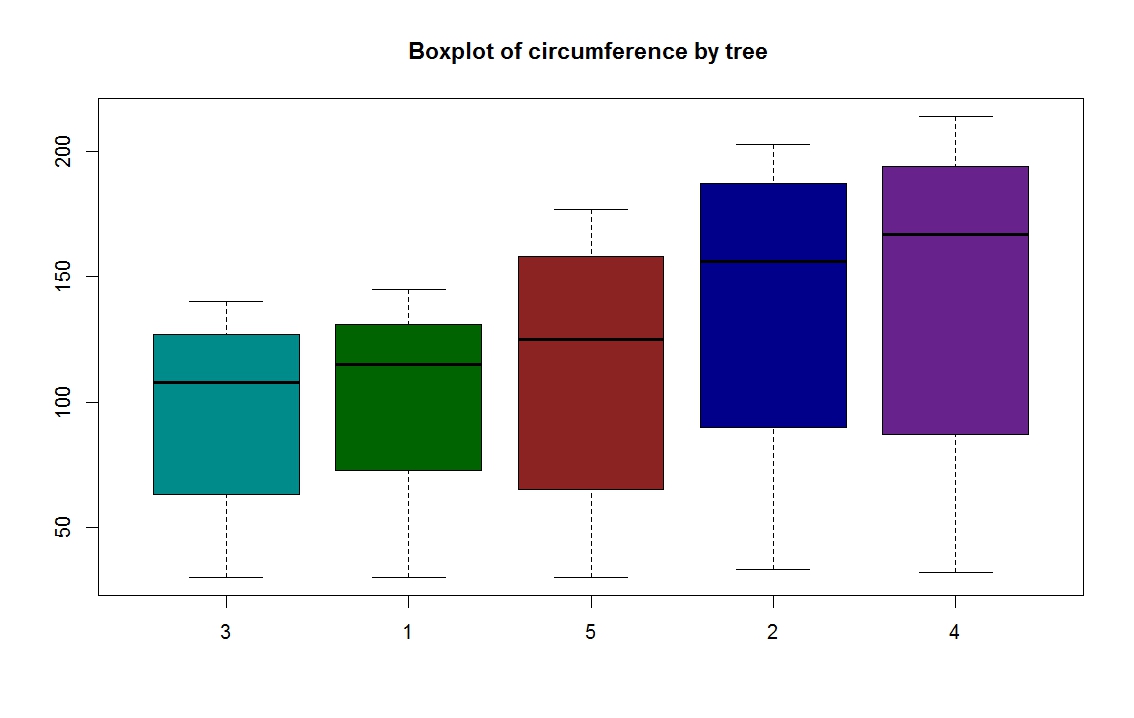
RG # 5: Grouped box plot
# data orage is from R library
data(Orange)
# to summarize the data
summary (Orange)
Tree age circumference
3:7 Min. : 118.0 Min. : 30.0
1:7 1st Qu.: 484.0 1st Qu.: 65.5
5:7 Median :1004.0 Median :115.0
2:7 Mean : 922.1 Mean :115.9
4:7 3rd Qu.:1372.0 3rd Qu.:161.5
Max. :1582.0 Max. :214.0
# defining own colors, for each group
mycol = c("darkcyan", "darkgreen", "brown4 ", "darkblue", "darkorchid4")
boxplot(circumference ~ Tree, data = Orange, main="Boxplot of circumference by tree ", col = mycol)
Subscribe to:
Posts (Atom)